Recent peer-reviewed publications:
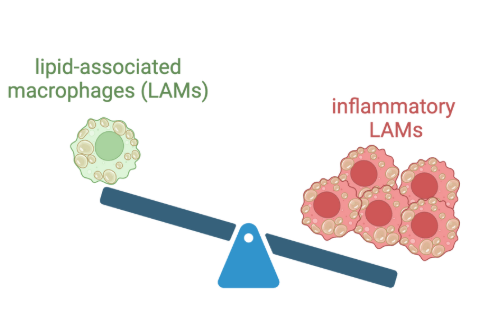
• Lapré TA, Hoeksema MA. Lipid associated macrophages as cellular markers of cardiometabolic disease.Cardiovasc Res. 2025 Nov 22:cvaf263. doi: 10.1093/cvr/cvaf263. Online ahead of print.
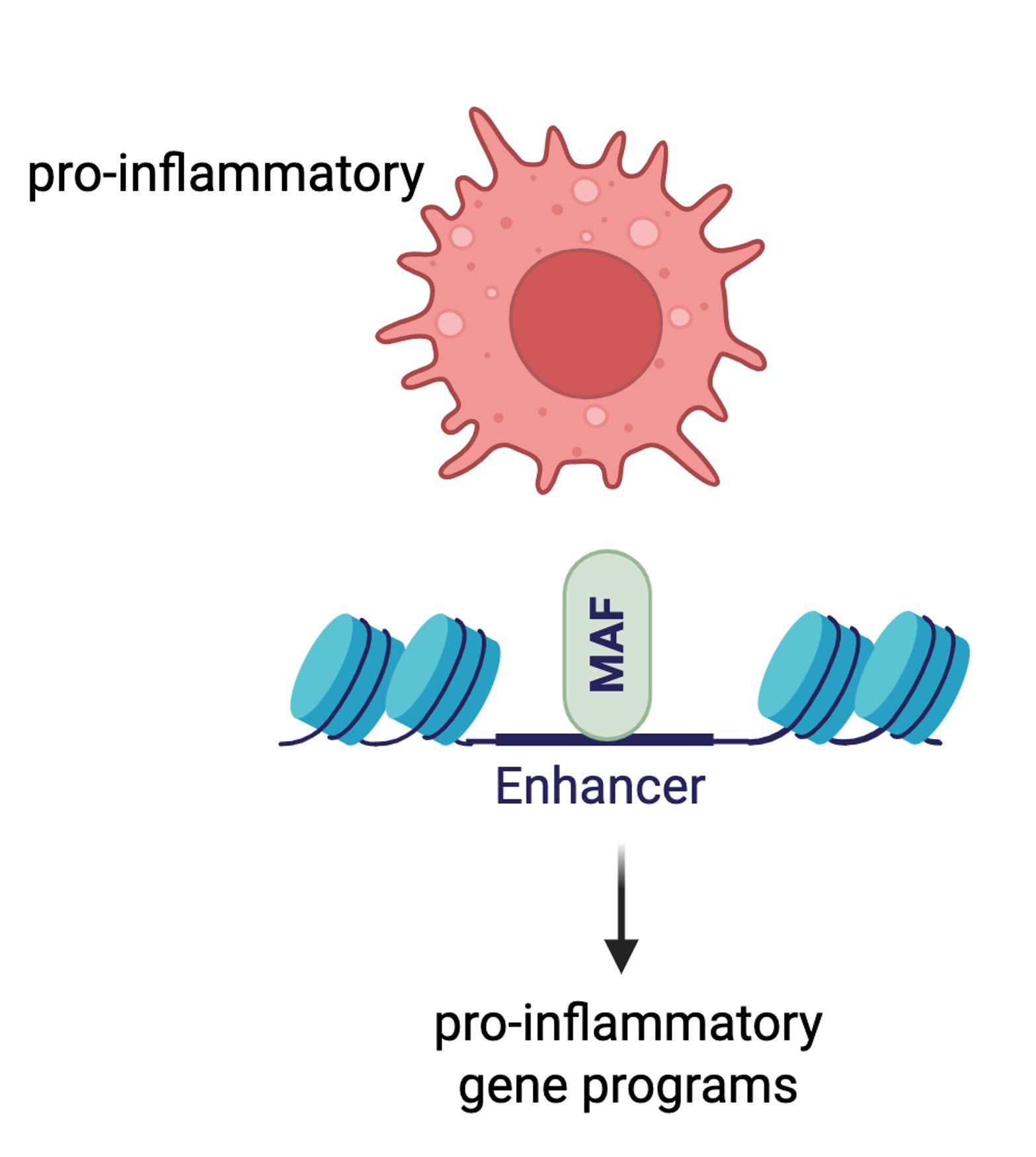
• Hoeksema MA. Fueling defense: PPARα enhances macrophage inflammatory responses. Cell Mol Immunol. 2025 Apr;22(4):461-462. doi: 10.1038/s41423-025-01265-y. Epub 2025 Mar 11.
• Reilly NA, Dekkers KF, Molenaar J, Arumugam S, Kuipers TB, Ariyurek Y, Hoeksema MA, Jukema JW, Heijmans BT. EPA Induces an Anti-Inflammatory Transcriptome in T Cells, Implicating a Triglyceride-Independent Pathway in Cardiovascular Risk Reduction. JACC Basic Transl Sci. 2025 Mar;10(3):383-395. doi: 10.1016/j.jacbts.2024.09.002. Epub 2024 Oct 30.
• Dinh B, Hoeksema MA, Spann NJ, Rendler J, Cobo I, Glass CK, Yeang C. Isolation and Cryopreservation of Highly Viable Human Peripheral Blood Mononuclear Cells From Whole Blood: A Guide for Beginners. J Vis Exp. 2024 Oct 25;(212). doi: 10.3791/66794.
• Lund SJ, Del Rosario PGB, Honda A, Caoili KJ, Hoeksema MA, Nizet V, Patras KA, Prince LS.Sialic Acid-Siglec-E Interactions Regulate the Response of Neonatal Macrophages to Group B Streptococcus. Immunohorizons. 2024 May 1;8(5):384-396. doi: 10.4049/immunohorizons.2300076.
• Reilly NA, Sonnet F, Dekkers KF, Kwekkeboom JC, Sinke L, Hilt S, Suleiman HM, Hoeksema MA, Mei H, van Zwet EW, Everts B, Ioan-Facsinay A, Jukema JW, Heijmans BT. Oleic acid triggers metabolic rewiring of T cells poising them for T helper 9 differentiation. iScience. 2024 Mar 12;27(4):109496. doi: 10.1016/j.isci.2024.109496. eCollection 2024 Apr 19.
• van Os BW, Vos WG, Bosmans LA, van Tiel CM, Toom MD, Beckers L, Admiraal M, Hoeksema MA, de Winther MP, Lutgens E. CD40L modulates CD4+ T cell activation through receptor for activated C kinase 1.Eur J Immunol. 2023 Sep 8:e2350520. doi: 10.1002/eji.202350520.
• Czimmerer Z, Patsalos A, Hoeksema MA. Editorial: Transcriptional regulation of macrophage function. Front Immunol. 2023 Nov 2;14:1321064.
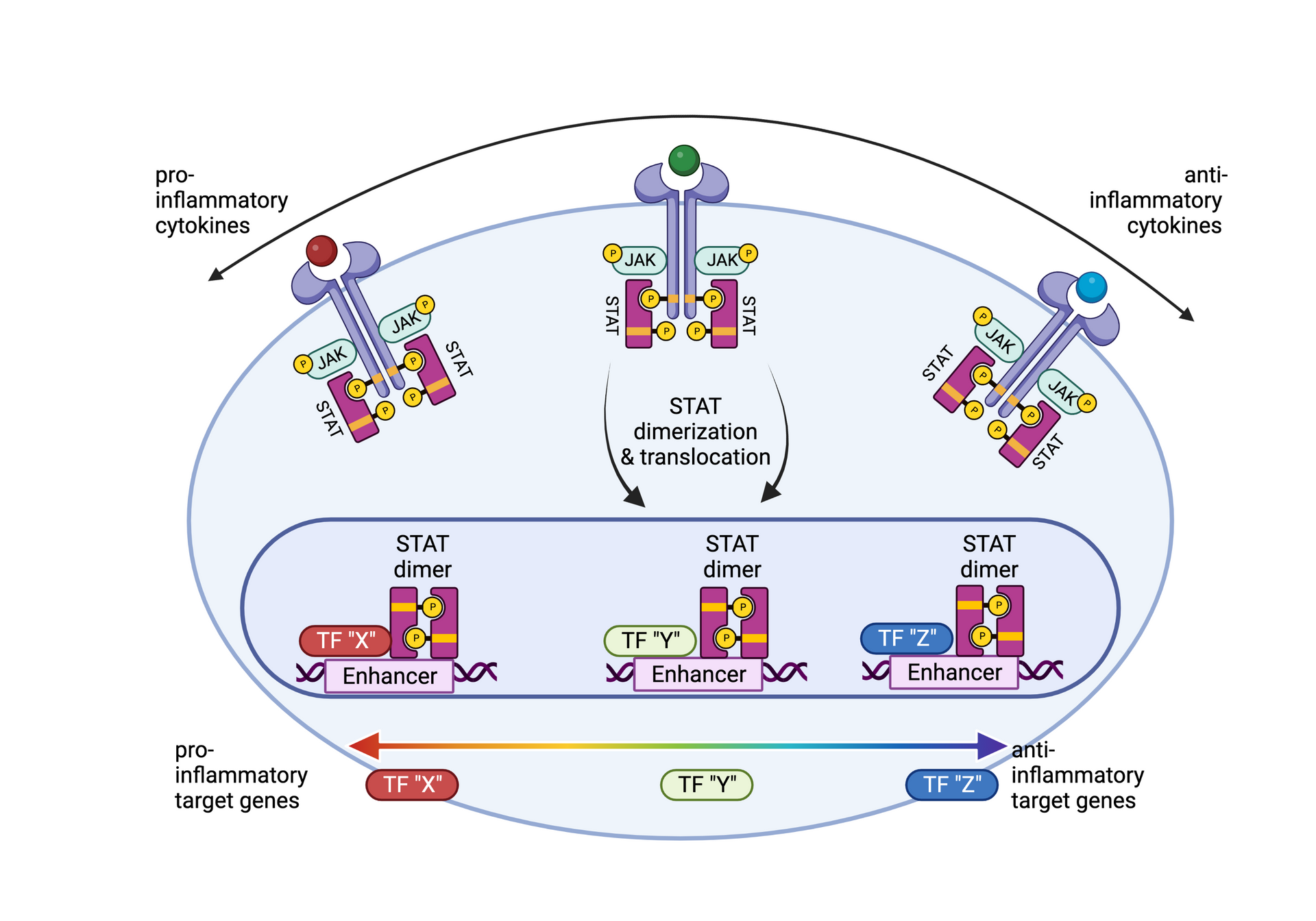
• Siebeler R, de Winther MPJ, Hoeksema MA. The regulatory landscape of macrophage interferon signaling in inflammation. J Allergy Clin Immunol. 2023 Aug;152(2):326-337. doi: 10.1016/j.jaci.2023.04.022. Epub 2023 Jun 3.
• van Wouw SAE, van den Berg M, El Ouraoui M, Meurs A, Kingma J, Ottenhoff R, Loix M, Hoeksema MA, Prange K, Pasterkamp G, Hendriks JJA, Bogie JFJ, van Klinken JB, Vaz FM, Jongejan A, de Winther MPJ, Zelcer N. Sterol-regulated transmembrane protein TMEM86a couples LXR signaling to regulation of lysoplasmalogens in macrophages. J Lipid Res. 2023 Feb;64(2):100325. doi: 10.1016/j.jlr.2022.100325.
• Czimmerer Z, Halasz L, Daniel B, Varga Z, Bene K, Domokos A, Hoeksema M, Shen Z, Berger WK, Cseh T, Jambrovics K, Kolostyak Z, Fenyvesi F, Varadi J, Poliska S, Hajas G, Szatmari I, Glass CK, Bacsi A, Nagy L. The epigenetic state of IL-4-polarized macrophages enables inflammatory cistromic expansion and extended synergistic response to TLR ligands. Immunity. 2022 Nov 8;55(11):2006-2026.e6. doi: 10.1016/j.immuni.2022.10.004.
• Breuss MW, Yang X, Schlachetzki JCM, Antaki D, Lana AJ, Xu X, Chung C, Chai G, Stanley V, Song Q, Newmeyer TF, Nguyen A, O'Brien S, Hoeksema MA, Cao B, Nott A, McEvoy-Venneri J, Pasillas MP, Barton ST, Copeland BR, Nahas S, Van Der Kraan L, Ding Y; NIMH Brain Somatic Mosaicism Network, Glass CK, Gleeson JG. Somatic mosaicism reveals clonal distributions of neocortical development. Nature. 2022 Apr;604(7907):689-696.
• Honda A, Hoeksema MA, Sakai M, Lund SJ, Lakhdari O, Butcher LD, Rambaldo TC, Sekiya NM, Nasamran CA, Fisch KM, Sajti E, Glass CK, Prince LS. The Lung Microenvironment Instructs Gene Transcription in Neonatal and Adult Alveolar Macrophages. J Immunol. 2022 Apr 15;208(8):1947-1959. doi: 10.4049
• Willemsen L, Chen HJ, van Roomen CPAA, Griffith GR, Siebeler R, Neele AE, Kroon J, Hoeksema MA*, de Winther MPJ*. Monocyte and Macrophage Lipid Accumulation Results in Down-Regulated Type-I Interferon Responses. Front Cardiovasc Med. 2022 Feb 10;9:829877. *shared senior author
• Hoeksema MA, Shen Z, Holtman IR, Zheng A, Spann NJ, Cobo I, Gymrek M, Glass CK. Mechanisms underlying divergent responses of genetically distinct macrophages to IL-4. Sci Adv. 2021 Jun 16;7(25):eabf9808.
• Shen Z, Li RZ, Prohaska TA, Hoeksema MA, Spann NJ, Tao J, Fonseca GJ, Le T, Stolze LK, Sakai M, Romanoski CE, Glass CK. Systematic analysis of naturally occurring insertions and deletions that alter transcription factor spacing identifies tolerant and sensitive transcription factor pairs. Elife. 2022 Jan 20;11:e70878.
• Weiss R, Spahn P, Chiang A, Liu Q, Li J, Hamill K, Rother S, Clausen T, Hoeksema MA, Timm B, Godula K, Glass CK, Tor Y, Gordts P, Lewis N, Esko J. Genome-wide screens uncover KDM2B as a modifier of protein binding to heparan sulfate. Nat. Chem. Biol. 2021 Jun;17(6):684-692.
• Neele AE, Chen HJ, Gijbels MJJ, van der Velden S, Hoeksema MA, Boshuizen MCS, Van den Bossche J, Tool AT, Matlung HL, van den Berg TK, Lutgens E, de Winther MPJ. Myeloid Ezh2 Deficiency Limits Atherosclerosis Development. Front Immunol. 2021 Jan 26;11:594603.
• Shen Z, Hoeksema MA, Ouyang Z, Benner C, Glass CK. MAGGIE: leveraging genetic variation to identify DNA sequence motifs mediating transcription factor binding and function. Bioinformatics. 2020 Jul 1;36(Suppl_1):i84-i92.